Cell line development is the project of cell engineering to investigate the mechanisms of biological functions through enhancing or suppressing the activities of biomolecules, typically proteins. Comparing overexpression, knock-in, knock-out, or knock-down cell lines with wild-type cells provides direct evidence to elucidate the roles of these biomolecules.
Besides overexpression, knock-in, knock-out, and knock-down cell lines, reporter cell line is also a powerful tool in biological research. These cells express markers like fluorescent proteins (GFP, YFP, mCherry) or luciferase, allowing researchers to track cell differentiation, identify tumor subpopulations, study cell–cell interactions, and perform in vivo imaging.
Ascent Research are expert in various techniques of cell engineering, such as gene editing systems (CRISPR/Cas9, TALEN), gene expression regulation systems (Tet-on/off, Cre-LoxP), gene delivery systems (lentiviral vectors, AAV, Adenoviral vectors), RNA-based gene silencing/modulation systems (microRNA mimics or inhibitors), and protein tagging.
Knock-in and Overexpression
Integration type: random insertion, site-specific insertion (safe harbor), and gene tagging (at either C-terminus or N-terminus).
Insertion fragment: SNP/point-mutation, tags (e.g. Flag, MiniSOG), reporter Gene (GFP, RFP, mCherry, Luciferease), and any gene of interest.
Host cell: Immune cells, stem cells, iPSCs, primary cells, and common tumor cell lines.
Delivery method (vector or RNP complex): Electroporation and lipofection.
Knock-out
Method: CRISPR RNP
Host cell: Immune cells, stem cells, iPSCs, primary cells, and common tumor cell lines.
Cell populations: Pool, polyclone, and single clone.
Other service: sgRNA synthesis
Note: For essential genes, knockdown is recommended instead of knockout. Please contact us for more details about engnieering strategies.
The following workflow presents the overview of cell line development using the lentiviral system:
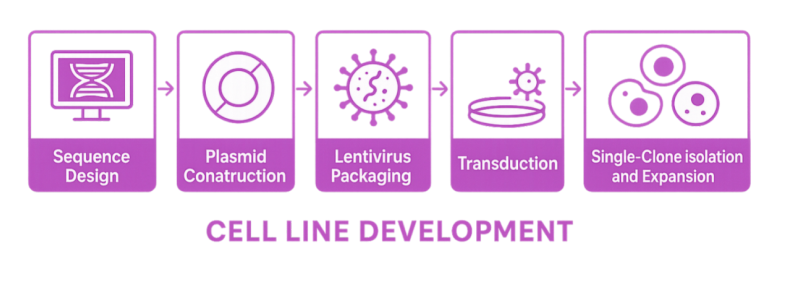
Sequence Design
Careful sequence design is critical for successful cell engineering. This process starts with obtaining the cDNA sequence of the target gene or protein from reliable sources such as BLAST (NCBI) and UniProt. Next, codon optimization is performed to enhance translation efficiency in the chosen cell line. This is followed by the selection of an appropriate plasmid backbone, taking into account factors like promoter strength, selection markers, and host compatibility. Restriction sites, linkers, or tags are also considered as elements incorporated into the design to facilitate cloning and downstream applications. Together, these steps form a comprehensive strategy for gene synthesis and vector construction.
Plasmid Construction
After sequence design, the next crucial phase is plasmid construction, which brings the designed sequence into a workable form for cell engineering. This process begins with obtaining the insert fragment(s), either through gene synthesis or PCR amplification from genomic cDNA. Meanwhile, the chosen plasmid backbone is linearized to prepare for insertion. Using seamless cloning or restriction enzyme cloning, the insert is integrated into the plasmid vector. Successful assembly is then verified by gel electrophoresis to confirm fragment sizes, followed by Sanger sequencing to validate the accuracy and integrity of the construct. This workflow ensures that the designed gene is precisely incorporated into a vector ready for downstream applications.
Viral Packaging
The lentiviral system is widely used for gene delivery into cells, especially when long-term and stable gene expression is needed. In the third generation lentiviral packaging system, the transfer plasmid carrying the gene of interest (GOI) is co-transfected with packaging plasmids and envelope plasmids into HEK293T cells. After 48 hours of incubation, the supernatant is collected, centrifuged to remove cell debris, and then filtered to ensure sterility. To increase viral concentration, polyethylene glycol (PEG) precipitation is employed. The titer of lentiviral particles is determined by qPCR. If further purification is required, sucrose gradient ultracentrifugation can be used to obtain ultra-purified lentiviral particles for downstream applications such as transduction of primary cells or in vivo delivery. Our lentiviral system is suitable for packaging gene overexpression vectors, chimeric antigen receptor (CAR) expression constructs, guide RNA expression vectors, and short hairpin RNA expression vectors with reporter genes.
Transduction
Transduction (or viral-mediated gene delivery) is the critical step following lentiviral packaging. Engineered viral particles infect target cells and deliver GOI into the host genome. Unlike transient transfection, transduction leverages the natural ability of viruses to efficiently introduce genetic material into various cells, including both dividing and non-dividing cells. This process ensures stable genomic integration of the transgene and long-term expression or modification.
Antibiotic Resistance Selection
Following transduction, antibiotic resistance selection is the next step, used to eliminate untransduced cells. This process relies on the inclusion of a selection marker (e.g., puromycin resistance, neomycin resistance), encoded within the viral vector. A killing curve is first established by treating parental cells with varying antibiotic concentrations to determine the minimum dose that kills >95% of cells within 3–7 days. This optimized dose is then applied to transduced cells, ensuring that only cells harboring integrated resistance genes survive, forming a pooled population of stably engineered cells.
Single-Clone Isolation and Expansion
Single-clone isolation is achieved through limiting dilution, typically plating cells at 0.5 cell/well or lower in a 96-well plate to statistically ensure clonality. After plating, wells are carefully monitored to confirm the presence of a single cell. Once single clones begin to expand, they are gradually transferred to larger culture vessels for further growth.
Once a stable cell line has been established, thorough quality control (QC) is essential to ensure consistency, functionality, and reliability. This involves confirmation of transgene integration, expression, and genetic modification, typically assessed by genomic PCR, qPCR, or RT-PCR. For protein-coding genes, methods such as western blotting, flow cytometry, or fluorescence imaging (if a reporter is included) may be used to evaluate protein expression levels and localization.
When cells are grown to the required amount, they are cryopreserved in vials containing an appropriate freezing medium and stored in liquid nitrogen for long-term stability. Prior to delivery, we prepare detailed documents with all quality control results and instructions for handling. The frozen vials are shipped using dry ice and insulated packaging to ensure cell viability upon arrival.
With these measures in place, researchers receive ready-to-use, validated stable cell lines that are consistent, traceable, and optimized for immediate application in the experiments.
We are familar with developing stable cell lines using the following host cells:
HeLa: robust and fast-growing, used for high-throughput screening and transfection studies.
HEK293: highly transfectable human cells, widely used for recombinant protein and viral vector production.
293T: high plasmid replication and protein expression levels.
A549: lung epithelial origin with stable growth and metabolism, useful for respiratory and drug response studies.
HepG2: retains key hepatic metabolic functions, useful as a standard host for liver metabolism and toxicity assays.
HCT 116: genetically well-characterized colorectal cancer cells, used for gene editing and signaling pathway research.
THP-1: human monocytic lineage with consistent differentiation potential, used for immune and inflammation modeling.
RAW 264.7: murine macrophage line with strong innate immune responses, commonly used for cytokine and phagocytosis studies.
4T1: highly tumorigenic and metastatic mouse breast cancer cells, suitable for in vivo-relevant cancer modeling.
ZR-75-1: hormone-responsive breast cancer cells, useful for estrogen receptor signaling and endocrine therapy research.
B16-F10: aggressive melanoma cells with predictable metastasis behavior, widely used in tumor biology studies.
MDA-MB-231: highly invasive triple-negative breast cancer cells (TNBC cells), a standard model for metastasis and drug resistance research.
We also have experience working on other primary cells and cell lines. Please contact us for further discussion.
Please contact us with a brief description of your project or specific need. One of our experts will get in touch with you soon to discuss your requirements for free.
